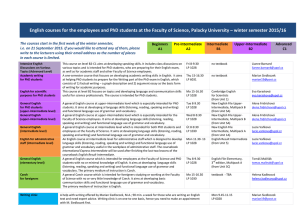
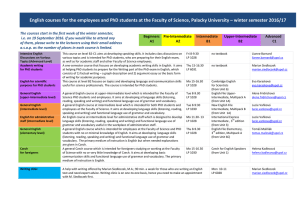
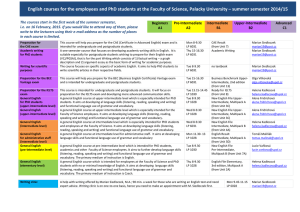
Sequence
Anuncio

First 2 days: Introduction
Aplicações biomédicas em plataformas
computacionais de alto desempenho
Aceleración de aplicaciones biomédicas sobre plataformas gráficas de altas prestaciones
Oswaldo Trelles
ortrelles@uma.es
PROGRAMA CAPES/DGU EDITAL No 040/2012
PROGRAMA HISPANO-BRASILEÑO DE COOPERACION INTERUNIVERSITARIA
LNCC, Petrópolis- Brasil, 2013
O.Trelles, PhD
Contents
Day
1
2
Contents description
Course presentation (15mins)
General overview
DNA sequencing / Assembly / Annotation
Sequence analysis concepts
Algorithms' internals: Blast
MuMer, HSPs identification
Introdution to Gene Expression data analysis
Sequential code internals
Speaker
ATR. Vasconcellos
Oswaldo Trelles
Oswaldo Trelles
This document provides an overview of the Introductory session (first two days)
O.Trelles, PhD
Contents: headlines
Headlines
Contents presentation
Basic concepts: biology, bioinformatics, HPC
Sequence analysis internals
Hands-on: sequential code
O.Trelles, PhD
Basic concepts on Biology
•
•
•
•
•
•
•
All living organisms are made-up of cells
Each cell contains the full genetic material (genome DNA sequence of {A, C, G, T})
The genome is organised in chromosomes
Chromosomes contains genes
Genes are the instructions to synthetize proteins (Genetic code)
The amount of each protein is regulated as response to changes in the environmental.
Metabolism that can involve dozens or hundreds of catalysed reactions in pathways.
O.Trelles, PhD
DNA sequencing
The DNA is long linear string of nucleotides
Sequence, a partial or the full genome ( string of a given
length.
Sequencing the process of determining the exact order of the
letters in the sequence.
Assembly: re-build up the original sequence from the “reads”
Solutions: an open issue
De-novo: a new genme is sequenced
Mapping: for re-sequenced genomes
Copy Number Variations, SNPs, etc make the problem still
more interesting ---if possible---
O.Trelles, PhD
Gene identification & function
Problems:
• Prokaryote & Eukaryote cells
• Intergenic regions.
• Coding regions
• Small portion of
genome
• Exons and introns.
• Conservation (mutations)
• Transposons and repeats
• Alternative splaicing
En color rosa se muestran los exones codificantes de la secuencia "prostate-specific antigen
promoter RT isolated from a patient with prostate cancer"
O.Trelles, PhD
Functional annotation
Biological sequence annotation is the process of finding, recovering and incorporating
relevant biological information available in public databases in relation to an individual
or massive collection of sequences.
New insights about function, cellular
location, phylogeny, biological process
and/or protein structure, etc.
In general is the next step in genome and
EST sequencing
O.Trelles, PhD
Transcriptomics
Transcripts (RNA) data
Genes modify their expression levels as response to
environmental stimuli, tissue location, time course...
Variations in gene expression patterns can lead
profound effects on biological functions being at the
core of altered physiologic and pathologic
processes.
Large scale technologies are changing our view of
the biological processes, including their dynamics.
Identify genes that share expression patterns and
hence might be regulated together are assumed to
be in the same genetic pathway.
O.Trelles, PhD
Gene-expression data analysis
Error removal: for reproducibility, reliability, compatibility and standardization
of data
Differential expression. identify over/under expressed
The gene “expression profile” represents the different levels of
expression along different experiments.
Each gene has its own particular expression profile, but, it can be
quite similar to the profile in other genes
Clustering: identify genes that share a similar expression
profile
distance measure (Euclidean, correlation, ...)
Method to proceed (hierarchical, kmeans, partitional, etc).
O.Trelles, PhD
Basic concepts on Bioinformatics
Bioinformatics:
Computer sciences applied to the processing of biological data
Different areas associated to the different type of data
Identificación de genes
Protein Sequences
Sequence Comparison
Clonación
Estadística comparativa
secuenciación
Rutas metabólicas
Sequence / structure function
filogenia
Databases
New technologies
Seq DNA Estructura DNA
Modelado
Molecular
Comparación de
estructuras
Statistical analysis
Protein Seq protein Structure
Computer programming
Estudios evolutivos
Expresión génica
Web servers
Integración de servicios
Metabolites
O.Trelles, PhD
Basis of parallel programing
•
•
•
•
•
Parallel architecture taxonomy
Parallel programming models
Sources of ineficiency in parallel programming
Performance measurements
Hands-on: bioinformatics application’s internals
O.Trelles, PhD
Sequence Analysis Internals
The essentials of biological sequence analysis
towards its computational aspects
Formal definition:
A sequence is a string f characters representing
DNA nucleotides or the protein amino acids
DNA: A= {a,c,g,t|u}
Why to compare sequences
How to compare sequences
Computational aspects
qNEW-SEQ
KNOWN-SEQ
--SARGDFLNAA YALFFMRSHN FGHSDVLPVL
||||||||
||| |||||
|||||||
MMSARGDFLN-- YALSLMRSHN DEHSDVLPVL
qNEW-SEQ
–-CSLKHVAY WDAYQALIYW IKAMNQQTDTSI
||||||||
|||||| | |||||||||
KNOWN-SEQ DVCSLKHVAY –VFQALIYW IKAMNQQTTLDT
qNEW-SEQ
--RPPDDQAF GHHHLPQAMH --SRLYVPS-SK
|||
|
|| |
||||||| ||
KNOWN-SEQ TIRPPA---- GAFGLPTANT CISRLYVPSMSK
O.Trelles, PhD
Hands-on: sequential codes (1)
●
●
●
Kmers frequencies
Codon usage
Qnormalization
Kmers analysis: Sequential pseudocode (example)
int main(int ac, char** av){
checkParameters(file, K);
seq = malloc(SEQSIZE);
freq= calloc(pow(4,K));
f
= fopen(In.file,"rt");
Tot =readSeq(f,seq); // Load Seq into memory
fclose(f);
for(i=0;i<Tot-(K-1);i++){
n=kmerIndex(seq,i,K);
if(n!=-1)
freq[n]++;
}
printKmerFreqs(freq,K);
}
O.Trelles, PhD
Hands-on: sequential codes (1)
Qnormalization
O.Trelles, PhD