Jorge A. Bevilacqua1,6, Yves Mathieu2, MarOn Krahn2, Marc
Anuncio
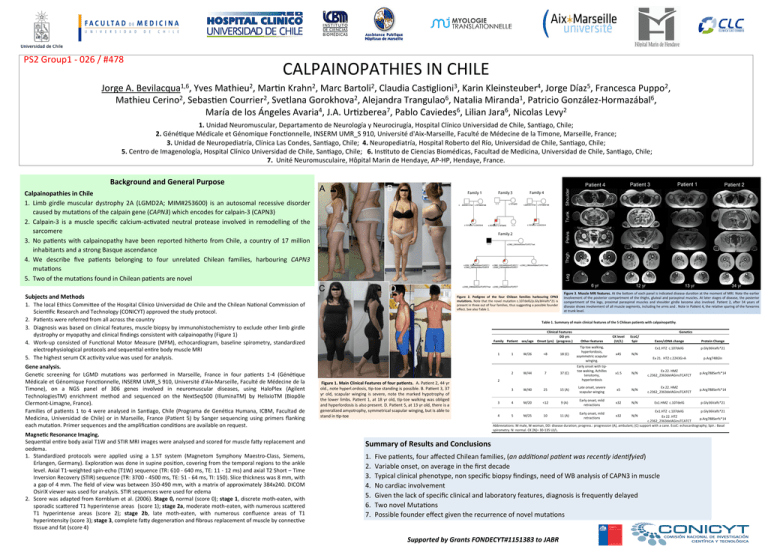
PS2Group1-026/#478 CALPAINOPATHIESINCHILE JorgeA.Bevilacqua1,6,YvesMathieu2,MarFnKrahn2,MarcBartoli2,ClaudiaCasFglioni3,KarinKleinsteuber4,JorgeDíaz5,FrancescaPuppo2, MathieuCerino2,SebasFenCourrier2,SvetlanaGorokhova2,AlejandraTrangulao6,NataliaMiranda1,PatricioGonzález-Hormazábal6, MaríadelosÁngelesAvaria4,J.A.UrFzberea7,PabloCaviedes6,LilianJara6,NicolasLevy2 1.UnidadNeuromuscular,DepartamentodeNeurologíayNeurocirugía,HospitalClínicoUniversidaddeChile,SanFago,Chile; 2.GénéFqueMédicaleetGénomiqueFoncFonnelle,INSERMUMR_S910,Universitéd'Aix-Marseille,FacultédeMédecinedelaTimone,Marseille,France; 3.UnidaddeNeuropediatría,ClínicaLasCondes,SanFago,Chile;4.Neuropediatría,HospitalRobertodelRío,UniversidaddeChile,SanFago,Chile; 5.CentrodeImagenología,HospitalClínicoUniversidaddeChile,SanFago,Chile;6.InsFtutodeCienciasBiomédicas,FacultaddeMedicina,UniversidaddeChile,SanFago,Chile; 7.UnitéNeuromusculaire,HôpitalMarindeHendaye,AP-HP,Hendaye,France. BackgroundandGeneralPurpose CalpainopathiesinChile 1. Limb girdle muscular dystrophy 2A (LGMD2A; MIM#253600) is an autosomal recessive disorder causedbymutaFonsofthecalpaingene(CAPN3)whichencodesforcalpain-3(CAPN3) 2. Calpain-3 is a muscle specific calcium-acFvated neutral protease involved in remodelling of the sarcomere 3. No paFents with calpainopathy have been reported hitherto from Chile, a country of 17 million inhabitantsandastrongBasqueascendance 4. We describe five paFents belonging to four unrelated Chilean families, harbouring CAPN3 mutaFons 5. TwoofthemutaFonsfoundinChileanpaFentsarenovel SubjectsandMethods 1. ThelocalEthicsCommiVeeoftheHospitalClínicoUniversidaddeChileandtheChileanNaFonalCommissionof ScienFficResearchandTechnology(CONICYT)approvedthestudyprotocol. 2. PaFentswerereferredfromallacrossthecountry 3. Diagnosiswasbasedonclinicalfeatures,musclebiopsybyimmunohistochemistrytoexcludeotherlimbgirdle dystrophyormyopathyandclinicalfindingsconsistentwithcalpainopathy(Figure1) 4. Work-up consisted of FuncFonal Motor Measure (MFM), echocardiogram, baseline spirometry, standardized electrophysiologicalprotocolsandsequenFalenFrebodymuscleMRI 5. ThehighestserumCKacFvityvaluewasusedforanalysis. Geneanalysis. GeneFc screening for LGMD mutaFons was performed in Marseille, France in four paFents 1-4 (GénéFque MédicaleetGénomiqueFoncFonnelle,INSERMUMR_S910,Universitéd'Aix-Marseille,FacultédeMédecinedela Timone), on a NGS panel of 306 genes involved in neuromuscular diseases, using HaloPlex (Agilent TechnologiesTM) enrichment method and sequenced on the NextSeq500 (IlluminaTM) by HelixioTM (Biopôle Clermont-Limagne,France). Families of paFents 1 to 4 were analysed in SanFago, Chile (Programa de GenéFca Humana, ICBM, Facultad de Medicina, Universidad de Chile) or in Marseille, France (PaFent 5) by Sanger sequencing using primers flanking eachmutaFon.PrimersequencesandtheamplificaFoncondiFonsareavailableonrequest. Magne<cResonanceImaging. SequenFalenFrebodyaxialT1WandSTIRMRIimageswereanalysedandscoredformusclefaVyreplacementand oedema. 1. Standardized protocols were applied using a 1.5T system (Magnetom Symphony Maestro-Class, Siemens, Erlangen,Germany).ExploraFonwasdoneinsupineposiFon,coveringfromthetemporalregionstotheankle level.AxialT1-weightedspin-echo(T1W)sequence(TR:610-640ms,TE:11-12ms)andaxialT2Short–Time InversionRecovery(STIR)sequence(TR:3700-4500ms,TE:51-64ms,TI:150).Slicethicknesswas8mm,with agapof4mm.Thefieldofviewwasbetween350-490mm,withamatrixofapproximately384x240.DICOM OsiriXviewerwasusedforanalysis.STIRsequenceswereusedforedema 2. ScorewasadaptedfromKornblumetal.(2006).Stage0,normal(score0);stage1,discretemoth-eaten,with sporadicscaVeredT1hyperintenseareas(score1);stage2a,moderatemoth-eaten,withnumerousscaVered T1 hyperintense areas (score 2); stage 2b, late moth-eaten, with numerous confluence areas of T1 hyperintensity(score3);stage3,completefaVydegeneraFonandfibrousreplacementofmusclebyconnecFve Fssueandfat(score4) Figure3.MuscleMRIfeatures.AttheboVomofeachpanelisindicateddiseaseduraFonatthemomentofMRI.Notetheearlier involvementoftheposteriorcompartmentofthethighs,glutealandparaspinalmuscles.Atlaterstagesofdisease,theposterior 2, ater 34 years of compartment of the legs, proximal paraspinal muscles and shoulder girdle become also involved. PaFent diseaseshowsinvolvementofallmusclesegments,includinghearmsand.NoteinPaFent4,therelaFvesparingoftheforearms attrunklevel. Figure 2. Pedigree of the four Chilean families harbouring CPN3 muta<ons.NotethatthenovelmutaFonc.107delG/p.Gly36Valfs*21is presentinthreeoutoffourfamilies,thussuggesFngapossiblefounder effect.SeealsoTable1. Table1.Summaryofmainclinicalfeaturesofthe5Chileanpatientswithcalpainopatthy. ClinicalFeatures DDyrs Family Patient sex/age Onset(yrs) (progress.) 1 Figure1.MainClinicalFeaturesoffourpa<ents.A.PaFent2,44yr old.,notehyperl.ordosis,Fp-toestandingispossible.B.PaFent3,37 yr old, scapular winging is severe, note the marked hypotrophy of thelowerlimbs.PaFent1,at18yrold,Fp-toewalkingwasobliged andhyperlordosisisalsopresent.D.PaFent5,at13yrold,thereisa generalizedamyotrophy,symmetricalscapularwinging,butisableto standinFp-toe 1 W/26 <8 Genetics Exon/cDNAchange ProteinChange Ex1.HTZc.107delG p.Gly36Valfs*21 Ex21.HTZc.2243G>A p.Arg748Gln 18(C) Tip-toewalking, hyperlordosis, asymmetricscapular winging. x45 N/N x1.5 N/N Ex22.HMZ c.2362_2363delAGinsTCATCT p.Arg788Serfs*14 2 W/44 7 37(C) Earlyonsetwithtiptoewaking,Achilles tenotomy, hyperlordosis 3 W/40 25 15(A) Lateonset,severe scapularwinging x5 N/N Ex22.HMZ c.2362_2363delAGinsTCATCT p.Arg788Serfs*14 4 M/20 <12 9(A) Earlyonset,mild retractions x32 N/N Ex1.HMZc.107delG p.Gly36Valfs*21 2 3 Otherfeatures CKlevel (UI/L) EcoC/ Spir Ex1.HTZc.107delG p.Gly36Valfs*21 4 5 M/25 10 11(A) x32 N/N Ex22.HTZ p.Arg788Serfs*14 c.2362_2363delAGinsTCATCT Abbreviations:Mmale,Wwoman,DD:diseaseduration;progress.:progression(A),ambulant;(C)supportwithacane.EcoC:echocardiography;Spir.:Basal spirometry.N:normal.CK[N]=30-135UI/L. Earlyonset,mild retractions SummaryofResultsandConclusions 1. 2. 3. 4. 5. 6. 7. FivepaFents,fouraffectedChileanfamilies,(anaddi+onalpa+entwasrecentlyiden+fyied) Variableonset,onaverageinthefirstdecade Typicalclinicalphenotype,nonspecificbiopsyfindings,needofWBanalysisofCAPN3inmuscle Nocardiacinvolvement Giventhelackofspecificclinicalandlaboratoryfeatures,diagnosisisfrequentlydelayed TwonovelMutaFons PossiblefoundereffectgiventherecurrenceofnovelmutaFons SupportedbyGrantsFONDECYT#1151383toJABR











